Neurodegenerative diseases (such as Alzheimer’s and Parkinson’s) that affect millions of people, are related by the aggregation of the peptide into nanoassemblies known as amyloid fibrils. Our research is focused on understanding the driving force for this aggregation and the design of small molecules to prevent it.
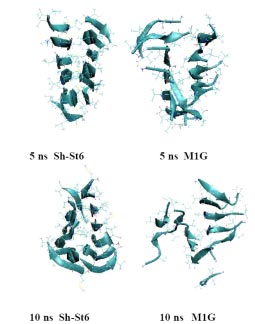
Supercomputers help us understand the atomistic details in the aggregation of the hexapeptide fragments of Tau, Insulin and Aβ peptides. We start with X-Ray crystal structure of each hexapeptide, select a decamer (ten adjacent molecules in the crystal lattice), and surround them by thousands of water molecules. The computer applies Newton’s laws to simulate evolution of this system, with and without small molecules present. Some of these molecules have been reported to inhibit protein aggregation. Alternatively, some amino acids in hexapeptides can be mutated. Elaborate analysis methods are used to observe the structural changes.
As a result of this analysis, we found which small molecules and mutations lead to the fastest disaggregation of the ten hexapeptides, as shown on the Figure. This knowledge could contribute to the rational design of more efficient amyloid inhibitors and amyloid-specific biomarkers for diagnostic purposes.
Qualitative Structure-Activity Relationships and virtual screening studies (including docking and molecular dynamic simulation) are being planned for further understanding of the structural requirements and for the discovery of the new inhibitor compounds.